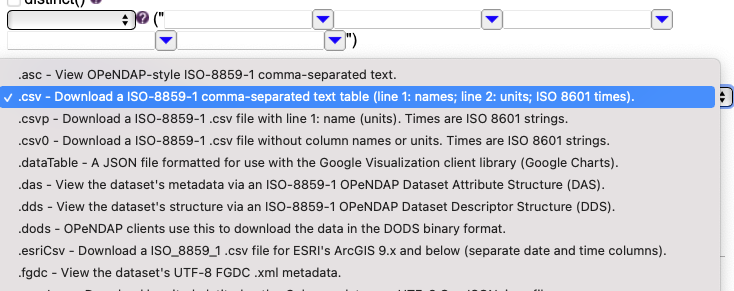
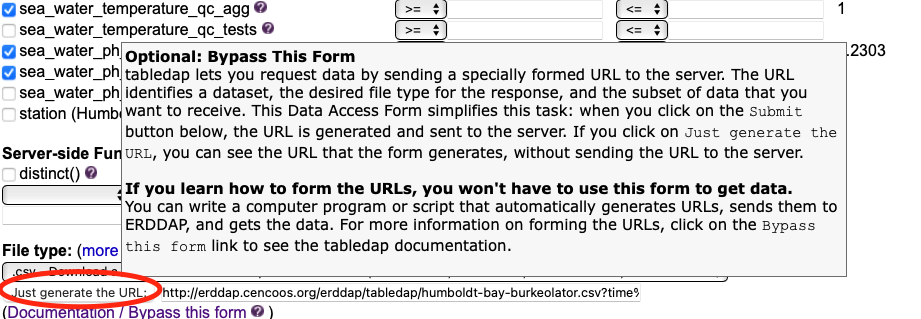
1. How to find the data¶
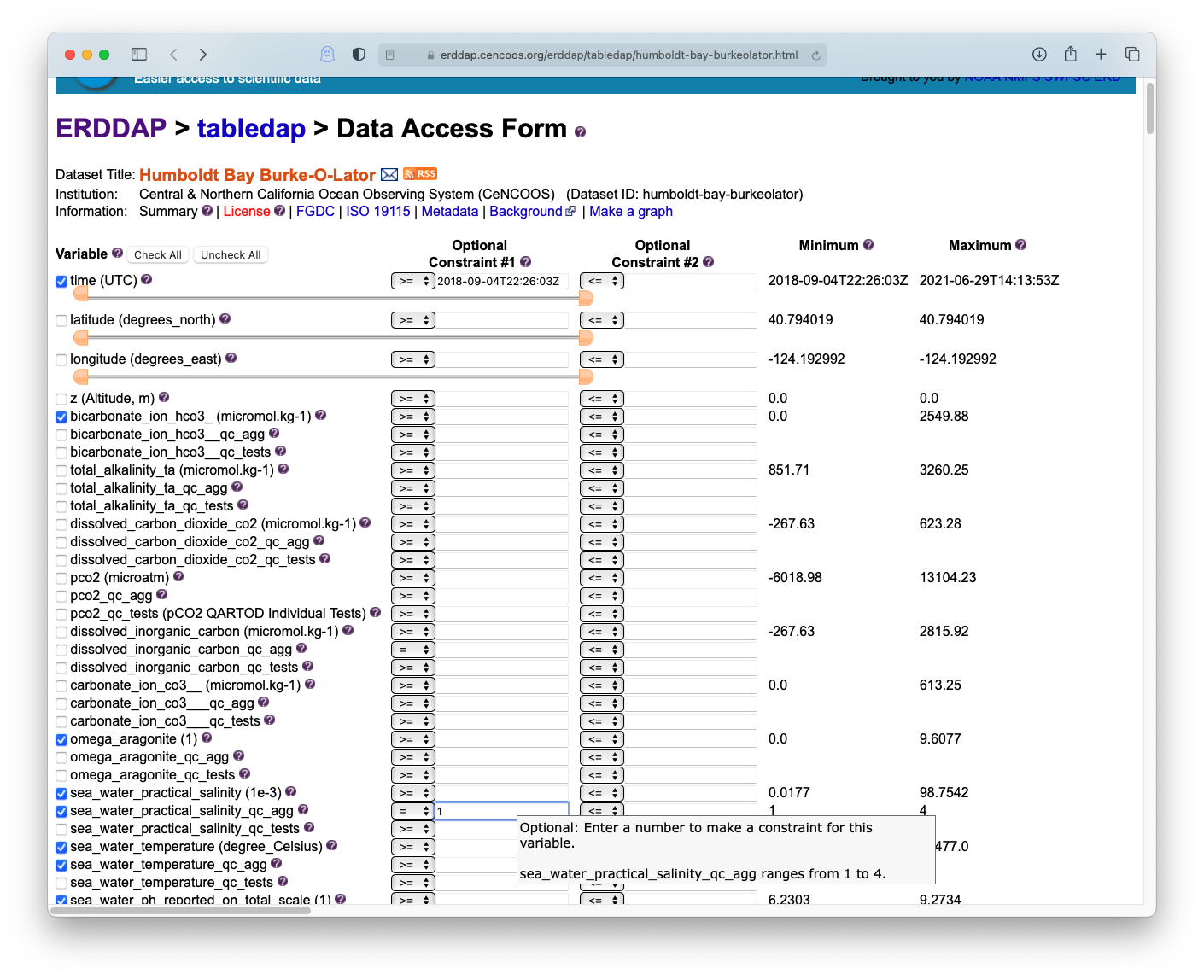
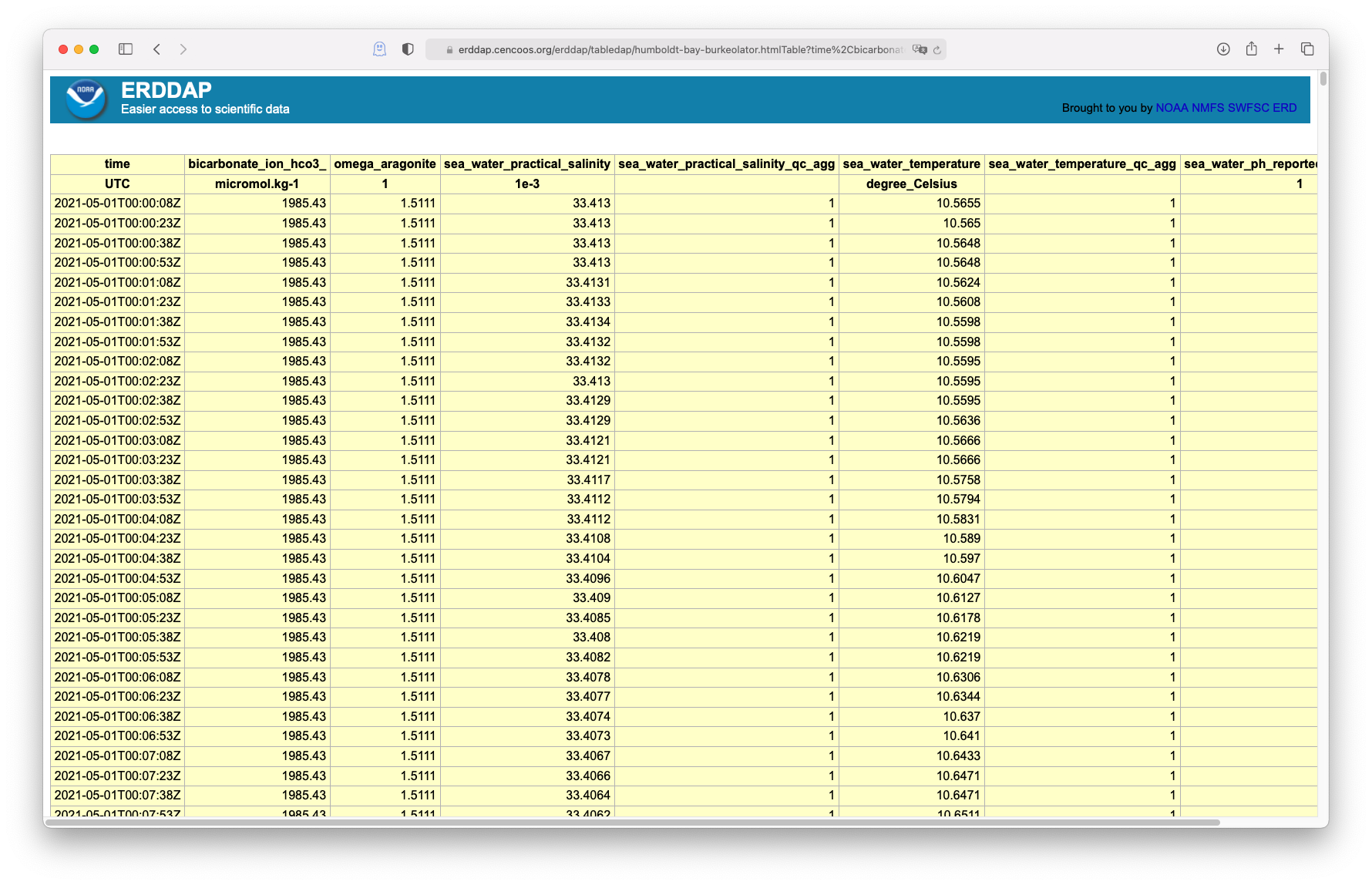
Try navigating to the CeNCOOS ERDDAP page: erddap.cencoos.org and searching for the station. The search is fairly elastic and will look through both the dataset title and id, as well as metadata which can include keywords.
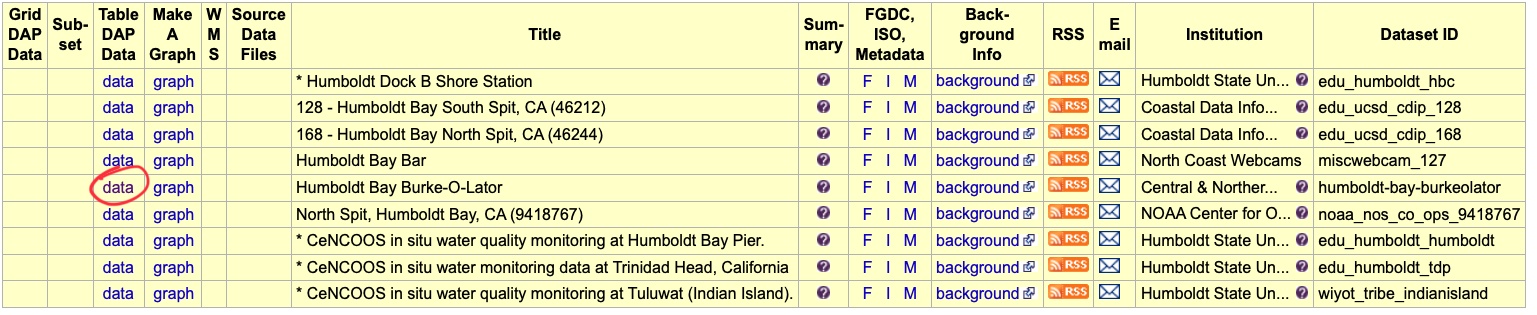
For example try searching for Humboldt or burkeolator